Abstract
The receptor tyrosine kinase FLT3 gene is commonly mutated in acute myeloid leukemia (AML) and its mutation is associated with significantly more aggressive disease and poor outcome. However, in the co-presence of NPM1 mutation, the disease is less aggressive and is associated with better outcome. FLT3 inhibitor midostaurin (Rydapt; Novartis Pharmaceuticals, Inc.) was recently approved for patients with FLT3 mutations. While the presence ofa FLT3 mutation is commonly associated with AML,it can develop in less acute myeloid neoplasms, such as MDS and CMML, and implies transformation into more acute phase. In this context, the FLT3 mutation is typically detected in a subclone of the original chronic myeloid clone. The prevalence of subclonal FLT3 mutations is unknown, as is the clinical relevance of subclonal FLT3, especially when treated with FLT3 inhibitors. We used variant allele frequency (VAF) and next generation sequencing (NGS) to determine if FLT3 mutations are founding or secondary mutations and to characterize the co-mutational background in large number of consecutive patients who presented with a clinical impression of myeloid neoplasm.
Methods: A total of 6390 consecutive bone marrow aspirate samples or peripheral blood samples were submitted with clinical impression of AML, MDS, or MPN between January 2014 and mid 2017 were tested for mutations in myeloid genes using NGS. We used the TruSight Myeloid panel (Illumina, San Diego, CA) for detecting missense mutations and fragment length analysis (FLA) for detecting internal tandem repeat (ITD) in FLT3 . DNA was extracted from samples using the QIAamp DNA Mini Kit. This NGS testing covers mutations in 54 myeloid-related genes. The average depth of sequencing was 10,000x.
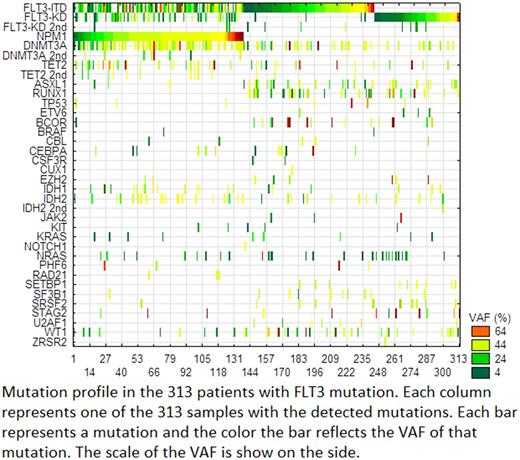
Results: A total of 2670 (42%) of the samples showed a mutation in one or more of the tested genes. Of the samples with mutations, FLT3 ITD and kinase domain (KD) mutations were detected in 313 (12%) patients. Of these patients, 208 (66.5%) had ITD and 117 (37.4%) had KD mutations. Twelve patients (3.8%) had both ITD and KD mutations. NPM1 mutation was detected in 138 (44%) of the patients. Most of FLT3 -KD mutations were in codon 835, but mutations in codons 836, 839, 842 and 845 were also found. Additionally, missense mutations in codons 576, 592, and 593 at the site of ITD were detected. To distinguish a FLT3 mutation as a founding mutation rather than a secondary mutation, we used a lower VAF of 10% as a cut-off. Using this approach, we found that 27% (115/313) of patients had secondary FLT3 mutations. Of these subclonal FLT3 mutants, NPM1 co-mutation was detected in 61 (53%) patients, but was not statistically significant (P=0.06). Patients with FLT3 mutation as a subclone had a significantly higher number of co-mutated genes (P-value <0.0001). This group of patients had higher (P≤0.01) mutations in TET2, DNMT3A, NRAS, IDH2, EZH2, and BCOR, consistent with an MDS/CMML background. On the other hand, patients with both FLT3 and NPM1 mutations had a significantly lower number of co-mutant genes (P<0.0001) and particularly lower (P≤0.01) mutation rate in U2AF1, SF3B1, SRSF2, RUNX1, IDH1, BCOR, and AXL1 .
Conclusion: FLT3 mutations are frequently (27%) a secondary mutation in a subclone of the neoplastic founding mutations. This appears to be more common when FLT3 mutation is developing in a background of MDS/CMML. The co-presence of an NPM1 mutation is more commonly associated with fewer co-mutations in MDS-related genes. This data suggests that in addition to detecting ITD by FLA, using NGS for profiling mutations in FLT3 -KD and other myeloid genes should be considered whenever FLT3 mutations are evaluated. Further, the clinical decision for combination therapy should consider the entire mutation profile and not only mutations in FLT3 .
Ma: NeoGenomics: Employment. De Dios: NeoGenomics: Employment. Funari: NeoGenomics: Employment. Sudarsanam: NeoGenomics: Employment. Jiang: NeoGenomics: Employment. Agersborg: NeoGenomics: Employment. Hummel: NeoGenomics: Employment. Blocker: NeoGenomics: Employment. Albitar: NeoGenomics: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal